- SybrGreen : assymetric cyanine that inserts itself into the double strand DNA little groove. When inserted, it emits a fluorescence at 525nm wavelength, usable at FAM channel in a PCR platform. SybrGreen is the most used dye in non specific qPCR and have very strong fluorescence intensity. It gives two different types of information : Target quantity with curves correlated to calibration curves and amplicon quality with melting curves. This last type of curve is obtained by doing a additional amplification program to amplification one (pretty easy to setp tup, ask if you do not find one) and give, after data analysis, the amplicon Tm depending of its size and base composition. Every different amplicon will have a different Tm.

Resolight (Roche) or LC green (for HRM application) : HRM (for High Resolution Melting) is a technology using next generation of intercalating dye inserting more efficiently in the DNA groove than SybrGreen. Indeed, whereas SybrGreen insert itself every 5 to 8 bases among DNA groove, HRM dyes (like LC green) insert in contiguous way and leave no gaps in insertion. Thus, single base mutation or little sequence difference are detected more easily with a fully inserted dye. That is why genotyping has made so huge advance since this technology has appeared. A single mutation can be detected because it will modify the Tm. Used wavelength are around 480nm excitation and 520nm emission.
Boxto : molecule from assymetric cyanines family like BEBO, occurs like Sybr Green. I do not know the intercalating density of this molecule but seems to be similar to SybrGreen. What is the interest so?
Its absorption/emission spectra is not close from FAM one (480/530bm) but closer to HEX/VIC one (520/560nm). This is very interesting used with FAM grafted probes because it allows to do a target amplification with FAM and to check primer-dimer formation and amplicon type adding BOXTO to do a melting curve step. And because emission spectra are different enough, this will not give any problem for analysis considering overlapping. And it WORKS !!!
This molecule can be found at TATAA biocenter (http://www.tataa.com/webshop/Dyes/BOXTO/Detailed-product-flyer.html)
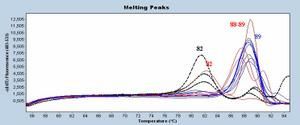
Hereunder a little assay made several months ago. One target but 4 differents intercalating dyes or kits comprising a specific dye :
- Boxto (red) : weak signal but it is normal considering that the analysis here is made with FAM channel instead of VIC normal one.
- Resolight (blue) : from Roche master mix
- MESA Green (black) : Sybr Green solution optimized by Eurogentec
- SybrGreen (green) : from Roche master mix

For next experiments, I will use Resolight more often and also Boxto from time to time with the ability to do melting curve with FAM target system amplified.
Waiting for that, have good curves and be bright...