Here is a very good publication for those of you interested by qPCR technique, fluorophores and quenching to improve your own detection system. A good synthesis of things to know when you want to optimize.
It pinpoints for example on the very important choice to do for fluorophores and quenchers for probes. Indeed, without detailing physical aspect of energy interaction, excited or stable state, energy tranfer and others, this publication deal with main aspects for qPCR probe to take into account.
This Marras et al (2006) publication talks about :
Quenching mode (FRET or contact)
Probes types (nearby, 5’-nucléase, molecular beacon, Strand-displacement)
Energy transfer efficiency
Fluorophores-quenchers interaction
Publication is written with the following information
:
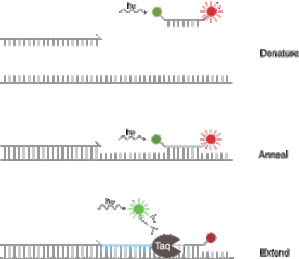
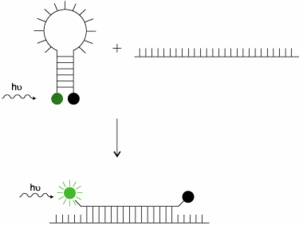
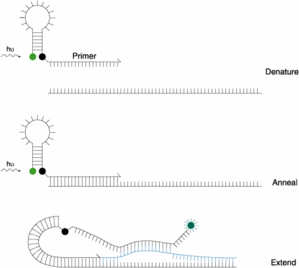
- Introduction define how to distinguish properly a double nearby probe (LC probes), an hydrolysis probe (TaqMan), a molecular beacon and a competitive hybridization probe (Yin-Yang)
- In chapter 3, energy transfer efficiency is detailed. It explains that a nearby quenching system is more efficient than a FRET one. But also that FRET system quenching, having a wider absorption spectrum, quenches more fluorophores than other quenching system.
- At last, many advices are given to do the best choice for the fluorophore/quencher couple considering qPCR device you use (Chapter 4). Here are few of them
* Considering fluorophore excitation mode (Laser, white light or DEL), excitation efficiency will be different for chose fluorophores. An Argon Laser with blue light will be less efficient for fluorophores having an optimum wavelenght over 540nm vs white light.
* If you have single target, you should use fluorophores having a wavelength between 495 and 540nm, that can detected by any qPCR device.
* If mulitplex qPCR, use fluorophores having the less overlapping spectra, even having a good qPCR device using efficient color compensation.
Another chapter, pretty interesting also, talks about fluorophore quantum yield (efficiency of a fluorophore to convert absorbed light into emitted light). Thus, a high yield gives better fluorescence intensity. But quantum yield can be modified by pH and temperature. Some fluorophore are more sensitive to temperature change. For example, ROX and HEX do not decrease fluorescence up to 85°C whereas FAM decreases steadily but in a moderate way up to 95°C. Cy5 can lose 70% of its quantum yield at 65°C. To offset this effect, probe concentration has to be increased.
Last but not least, it is recalled that a Guanosine base must never be as first probe base and being the closest to grafted fluorophore otherwise the natural quenching effect of G base will affect the fluorophore.
This publication has to be read by any qPCR user dealing with qPCR optimization and data analysis.
Enjoy or in french "Bonne lecture"
Reference
Selection of fluorophore and quencher pairs for fluorescent nucleic acid hybridization probes.
With
the introduction of simple and relatively inexpensive methods for
labeling nucleic acids with nonradioactive labels, doors have been
opened that enable
nucleic acid hybridization probes to be used for research and
development, as well as for clinical diagnostic applications. The use of
fluorescent hybridization probes that generate a
fluorescence signal only when they bind to their target enables
real-time monitoring of nucleic acid amplification assays. The use of
hybridization probes that bind to the amplification
products in real-time markedly improves the ability to obtain
quantitative results. Furthermore, real-time nucleic acid amplification
assays can be carried out in sealed tubes, eliminating
carryover contamination. Because fluorescent hybridization probes
are available in a wide range of colors, multiple hybridization probes,
each designed for the detection of a different nucleic
acid sequence and each labeled with a differently colored
fluorophore, can be added to the same nucleic acid amplification
reaction, enabling the development of high-throughput multiplex
assays. It is therefore important to carefully select the labels
of hybridization probes, based on the type of hybridization probe used
in the assay, the number of targets to be detected, and
the type of apparatus available to perform the assay. This chapter
outlines different aspects of choosing appropriate labels for the
different types of fluorescent hybridization probes used
with different types of spectrofluorometric thermal cyclers.